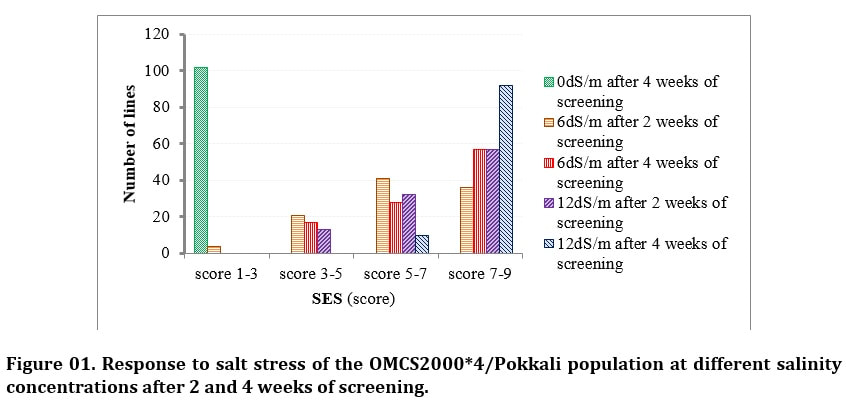
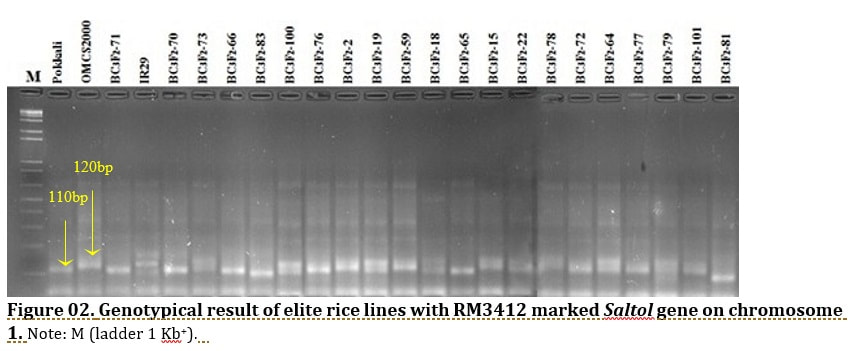
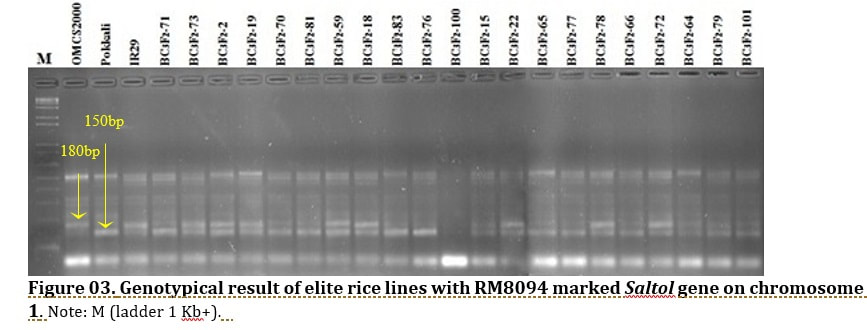
J. Biosci. Agric. Res. | Volume 19, Issue 01, 1576-1588| https://doi.org/10.18801/jbar.190119.192
Article type: Research article, Received: 15.07.2018, Revised: 24.12.2018, Date of Publication: 14 January 2019.
Article type: Research article, Received: 15.07.2018, Revised: 24.12.2018, Date of Publication: 14 January 2019.
Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage
Bui Phuoc Tam 1, Pham Thi Be Tu 1, Pham Van Ut 2, Pham Thu Dung 1, Phong Ngoc Hai Trieu 1, Nguyen Le Van 1, Tran Nhu Ngoc 1, Bien Anh Khoa 3
1 CuuLong Delta Rice Research Institute, Can Tho city, Vietnam
2 Department of Agriculture and Rural Development of Can Tho city, Can Tho city, Vietnam
3 Can Tho University, Can Tho city, Vietnam.
1 CuuLong Delta Rice Research Institute, Can Tho city, Vietnam
2 Department of Agriculture and Rural Development of Can Tho city, Can Tho city, Vietnam
3 Can Tho University, Can Tho city, Vietnam.
Abstract
Rice breeding of salinity tolerance based on a combination of phenotyping, genotyping and analyzing of genetic relationship was aiming to create the next generation with stable tolerance. In this study, 99 rice lines of the OMCS2000*4/Pokkali population in BC3F2 generation were screened in saline environments of 0, 6 and 12 dS m-1 at seedling stage. The tolerant lines were continuously tested the Saltol gene with 2 tightly markers, RM3412 and RM8094. The population was employed for the assessment of correlation, genetic variability, heritability and genetic advance for the related traits. Through phenotyping and genotyping, the population was tolerant to 6 dS m-1 for 4 weeks and 12 dS m-1 for 2 weeks, in which, 21/99 rice lines was highly tolerant and carried the Saltol gene. In this population, SES score and fresh biomass were the most significant traits and could explain over 70% of variability. SES scores were negatively correlated to almost all other traits, strongly to biomass, moderately to plant height, and lowly to root length. Analysis of variance revealed that sufficient genetic variation has been created for all traits. Relative magnitude of phenotypic coefficients of variation was higher than genotypic coefficients of variation for all the characters under study indicating environmental influence on the traits. High heritability coupled with high genetic advance was almost traits, exception of dry biomass. These indicated that direct selection can be effective for salinity tolerance improvement in the population under study.
Key Words: Salinity tolerance, Seedling stage, Genetic variation, Heritability and Genetic advance
Rice breeding of salinity tolerance based on a combination of phenotyping, genotyping and analyzing of genetic relationship was aiming to create the next generation with stable tolerance. In this study, 99 rice lines of the OMCS2000*4/Pokkali population in BC3F2 generation were screened in saline environments of 0, 6 and 12 dS m-1 at seedling stage. The tolerant lines were continuously tested the Saltol gene with 2 tightly markers, RM3412 and RM8094. The population was employed for the assessment of correlation, genetic variability, heritability and genetic advance for the related traits. Through phenotyping and genotyping, the population was tolerant to 6 dS m-1 for 4 weeks and 12 dS m-1 for 2 weeks, in which, 21/99 rice lines was highly tolerant and carried the Saltol gene. In this population, SES score and fresh biomass were the most significant traits and could explain over 70% of variability. SES scores were negatively correlated to almost all other traits, strongly to biomass, moderately to plant height, and lowly to root length. Analysis of variance revealed that sufficient genetic variation has been created for all traits. Relative magnitude of phenotypic coefficients of variation was higher than genotypic coefficients of variation for all the characters under study indicating environmental influence on the traits. High heritability coupled with high genetic advance was almost traits, exception of dry biomass. These indicated that direct selection can be effective for salinity tolerance improvement in the population under study.
Key Words: Salinity tolerance, Seedling stage, Genetic variation, Heritability and Genetic advance
Article Full-Text PDF
| 192.01.19.19_genetic_analysis_of_salinity_tolerance_in_a_rice_backcross_population_at_seedling_stage.pdf | |
| File Size: | 1263 kb |
| File Type: | |
Article Metrics
|
Share This Article
|
|
Article Citations
MLA
Tam B. P. et al. “Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage
.” Journal of Bioscience and Agriculture Research 19(01) (2019): 1576-1588.
APA
Tam, B. P., Tu, P. T. B., Ut, P. V., Dung, P. T., Trieu, P. N. H., Van, N. L., Ngoc, T. N., Khoa, B. A. (2019). Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage. Journal of Bioscience and Agriculture Research, 19(01), 1576-1588.
Chicago
Tam, B. P., Tu, P. T. B., Ut, P. V., Dung, P. T., Trieu, P. N. H., Van, N. L., Ngoc, T. N., Khoa, B. A. “Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage.” Journal of Bioscience and Agriculture Research 19(01) (2019): 1576-1588.
Harvard
Tam, B. P., Tu, P. T. B., Ut, P. V., Dung, P. T., Trieu, P. N. H., Van, N. L., Ngoc, T. N., Khoa, B. A. 2019. Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage. Journal of Bioscience and Agriculture Research, 19(01), pp. 1576-1588.
Vancouver
Tam, BP, Tu, PTB, Ut, PV, Dung, PT, Trieu, PNH, Van, NL, Ngoc, TN and Khoa, BA. Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage. Journal of Bioscience and Agriculture Research. 2019 January 19(01): 1576-1588.
Tam B. P. et al. “Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage
.” Journal of Bioscience and Agriculture Research 19(01) (2019): 1576-1588.
APA
Tam, B. P., Tu, P. T. B., Ut, P. V., Dung, P. T., Trieu, P. N. H., Van, N. L., Ngoc, T. N., Khoa, B. A. (2019). Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage. Journal of Bioscience and Agriculture Research, 19(01), 1576-1588.
Chicago
Tam, B. P., Tu, P. T. B., Ut, P. V., Dung, P. T., Trieu, P. N. H., Van, N. L., Ngoc, T. N., Khoa, B. A. “Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage.” Journal of Bioscience and Agriculture Research 19(01) (2019): 1576-1588.
Harvard
Tam, B. P., Tu, P. T. B., Ut, P. V., Dung, P. T., Trieu, P. N. H., Van, N. L., Ngoc, T. N., Khoa, B. A. 2019. Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage. Journal of Bioscience and Agriculture Research, 19(01), pp. 1576-1588.
Vancouver
Tam, BP, Tu, PTB, Ut, PV, Dung, PT, Trieu, PNH, Van, NL, Ngoc, TN and Khoa, BA. Genetic Analysis of Salinity Tolerance in a Rice Backcross Population at Seedling Stage. Journal of Bioscience and Agriculture Research. 2019 January 19(01): 1576-1588.
References
- Akhtar, S., Islam, M.M., Begum, S.N., Halder, J., Alam, M.K. and Manidas, A.C. (2010). Genetic analysis of f4 rice lines for salt tolerance at the reproductive stage. Progress. Agric. 21(1 & 2), 31–38.
- Allard, R.W. (1960). Principles of plant breeding. J. Wiley and Sons, London, pp 83-108.
- Bonilla, P.S., Dvorak, J., Mackill, D., Deal, K. and Gregorio, G. (2002). RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. The Philippines Agricultural Scientist, 85(1), 64-74.
- Bulmer, M.G. (1985). The mathematical theory of quantitative genetics. Clarendon Press, Oxford, U.K.
- Burton, G.W. and Devane, E.H. (1953). Estimating the heritability in tall fescue (Festuca arundinancea) from replicated clonal material. Agronomy J. 45, 478-481. https://doi.org/10.2134/agronj1953.00021962004500100005x
- Buu, B.C., Lang, N.T., Tao, P.B. and Bay, N.D. (1995). Rice breeding research strategy in the Mekong Delta. Proc. of the Int. Rice Res. Conf. Fragile Lives in Fragile Ecosystems, IRRI, Philippines, 739-755.
- De Ocampo M.P. (2014). Mapping of salinity tolerance in rice through genome-wide association mapping (GWAS). 4th International Rice Congress, Bangkok Thailand.
- DWR (Directorate of Water Resources), Ministry of Agriculture and Rural Development. (2018). Salinity intrusion in the Mekong River Delta (2015-2016), drought problem in Central Vietnam, Tay Nguyen, and solutions to overcome” (Vietnamse). http://www.tongcucthuyloi.gov.vn/Tin-tuc-Su-kien/Tin-tuc-su-kien-tong-hop/catid/12/item/2670/xam-nhap-man-vung-dong-bang-song-cuu-long--2015---2016---han-han-o-mien-trung--tay-nguyen-va-giai-phap-khac-phuc.
- Falconer, D.S. (1982). Introduction to quantitative genetics. Oliver and Boyd, London, p. 340.
- FAO (Food and Agriculture Organization) (2010). Report of salt affected agriculture, http://www. fao.org.
- Ferdose, J., Kawasaki, M., Taniguchi, M. and Miyake, H. (2009). Differential sensitivity of rice cultivars to salinity and its relation to ion accumulation and root tip structure. Plant Prod Sci. 12(4), 453–461. https://doi.org/10.1626/pps.12.453
- Fraga, T.I., Carmona, F.C., Anghinoni, I. and Marcolin, E. (2010). Attributes of irrigated rice and soil solution as affected by salinity levels of the water layer. R. Bras. Ci. Solo. 34,1049-1057. https://doi.org/10.1590/S0100-06832010000400005
- Gregorio G.B., Senadhira D. and Mendoza R.D. (1997). Screening rice for salinity tolerance. International Rice Research Institute discussion paper series No. 22, 1997.
- Gregorio, G.B. and Senadhira, D. (1993). Genetics analysis of salinity tolerance in rice. Theor. Appl. Genet. 86, 333-338. https://doi.org/10.1007/BF00222098
- Hakim, M.A., Juraimi, A.S., Hanafi, M.M., Ismail, M.R., Rafii, M.Y., Islam, M.M. and Selamat, A. (2014). The effect of salinity on growth, ion accumulation and yield of rice varieties. The Journal of Animal & Plant Sciences, 24(3), 874-885.
- Haq, T., Akhtar, J., Nawaz, S. and Ahmad, R. (2009). Morpho- physiological response of rice (Oryza sativa L.) varieties to salinity stress. Pak. J. Bot. 41 (6), 2943-2956.
- Hasanuzzaman, M., Fujita, M., Islam, M.N., Ahamed, K.U. and Nahar, K. (2009). Performance of four irrigated rice varieties under different levels of salinity stress. Int. J. Integ. Biol. 6, 85-90.
- Hospital, F. (2003). Marker-assisted breeding. In: Newbury HJ, editor. Plant molecular breeding. Oxford: Blackwell, 30–59.
- Hossain, F., Chandra Sarker, B., Kamaruzzaman, M., Abdul Halim, M., Najim Uddin, M. and Siddique, M. N. A. (2015). Physiological investigation of rice land races in a low temperature area of Bangladesh. Research Journal of Agriculture and Forestry Sciences, 3(7), 01-06.
- Hossain, M. A. and Siddique, M. N. A. (2015). Water-A limiting resource for sustainable agriculture in Bangladesh. EC Agriculture, 1(2), 124-137.
- Huyen, L.T.N., Cuc, L.M., Ham, L.H. and Khanh, T.D. (2013). Introgression the Saltol QTL into Q5BD, the elite variety of Vietnam using marker assisted selection (MAS). Am J Biosci. 1, 80-84. https://doi.org/10.11648/j.ajbio.20130104.15
- IRRI (International Rice Research Institute). (2006). Rice Knowledge Bank. www.knowledgebank.irri.org.
- IRRI (International Rice Research Institute). (2011). Laboratory Handbook of Molecular Marker Application for Rice Breeding. International Rice Research Institute.
- Islam, M. M., Adhikary, S. K., Gain, P., Rahman, M. M. and Siddique, M. N. A. (2004). Effect of plant growth regulators on callus induction and plant regeneration in another culture of rice. Pakistan J. Biol. Sci., 7, 331-334. https://doi.org/10.3923/pjbs.2004.331.334
- Ismail, A.M., Platten, J.D. and Miro, B. (2013). Physiological bases of tolerance of abiotic stresses in rice and mechanisms of adaptation. Oryza, 50(2), 91-99.
- Johnson, H.W., Robinson, H.F. and Comstock, R.E. (1955). Estimates of genetic and environmental variability in Soyabean. Agronomy J. 47, 314-318. https://doi.org/10.2134/agronj1955.00021962004700070009x
- Karim, D., Sarkar, U., Siddique, M. N. A., Miah, M. K. and Hasnat, M. Z. (2007). Variability and genetic parameter analysis in aromatic rice. Int. J. Sustain. Crop. Prod. 2(5), 15-18.
- Karim, D., Siddique, M. N. A., Sarkar, U., Hasnat, Z. and Sultana, J. (2014). Phenotypic and genotypic correlation co-efficient of quantitative characters and character association of aromatic rice. Journal of Bioscience and Agriculture Research, 1(1), 34-46.https://doi.org/10.18801/jbar.010114.05
- Lang, N.T., Buu, B.C. and Ismail, A.M. (2008). Molecular mapping and marker-assisted selection for salt tolerance in rice (Oryza sativa L.). Omonrice, 16, 50-56.
- Lang, N.T., Loc, L.Q., Ha, P.T.T., Tam, B.P. and Buu, B.C. (2017). Screening rice for salinity tolerance by phenotypic in population OM1490/Pokkali//OM1490 cross at seedling stage. International Journal of Current Innovation Research, 3 (7), 700-706.
- Lin, C.Y. (1978). Index selection for genetic improvement of quantitative characters. Theor. Appl. Genet. 52, 49–56. https://doi.org/10.1007/BF00281316
- Lutts, S., Kinet, J.M. and Bouharmont, J. (1995). Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J Exp Bot. 46, 1843–1852. https://doi.org/10.1093/jxb/46.12.1843
- Maclean, J.L., Dawe, D.C., Hardy, B. and Hettel, G.P. (2002). Rice Almanac. Los Banos (Philippines): International Rice Research Institute, pp.16-24.
- Maggio, A., Miyazaki, S., Veronese, P., Fujita T., Ibeas, J.I., Damsz, B., Narasimhan, M.L., Hasegawa, P.M., Joly, R.J. and Bressan, G.A. (2002). Does proline accumulation play an active role in stress-induced growth reduction? Plant J. 31, 699-712.https://doi.org/10.1046/j.1365-313X.2002.01389.x
- Mahmood, A., Latif, T. and Khan M.A. (2009). Effect of salinity on growth, yield and yield components in Basmati rice germplasm. Pak. J.Bot. 41(6), 3035-3045.
- Meena, B.L., Das, S.P., Meena, S.K., Kumari, R., Devi, A.G. and Devi, H.L. (2017). Assessment of GCV, PCV, Heritability and Genetic Advance for Yield and its Components in Field Pea (Pisum sativum L.). Int. J. Curr. Microbiol. App. Sci. 6(5), 1025-1033.https://doi.org/10.20546/ijcmas.2017.605.111
- Mishra, B., Singh, R.K. and Jetly, V. (1998). Inheritance pattern of salinity tolerance in rice. J. Gnet. & Breed. 52, 325-331.
- Mondal, S. and Borromeo, T.H. (2016). Screening of salinity tolerance of rice at early seedling stage. J. Biosci. Agric. Res. 10 (1), 843 - 847. https://doi.org/10.18801/jbar.100116.102
- Munns, R. (2009). Chapter 11: Strategies for Crop Improvement in Saline Soil. Book: Salinity and Water Stress: Improving Crop Efficiency, p. 101.
- Munns, R. and Tester, M. (2008). Mechanisms of salinity tolerance. Annual Review of Plant Biology, 59, 651-681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
- Negrão, S., Courtois, B., Ahmadi, N., Abreu, I., Saibo, N. and Oliveira, M.M. (2011). Recent updates on salinity stress in rice: from physiological to molecular responses. Book: Critical Reviews in Plant Sciences. Tayor & Francis. 30(4), 329-377.https://doi.org/10.1080/07352689.2011.587725
- Nicholas, F.W. (1987). Veterinary Genetics. Clarendon Press, Oxford.
- Platten, J.D., Egdane, J. and Ismail, A.M. (2013). Salinity tolerance, Na+ exclusion and allele mining of HKT1; 5 in Oryza sativa and O. glaberrima: many sources, many genes, one mechanism. BMC Plant Biol. 13-32.
- Ponnamperuma. F.N. (1994). Evaluation and improvement of lands for wetland rice production. In: (Eds). Senadhira, D. Rice and problem soils in South and Southeast Asia. IRRl, Manila, Philippines. Discussion Paper Series No.4. pp. 3-19
- Ray, P.K.S. and Islam, M.A. (2008). Genetic analysis of salinity tolerance in rice. Bangladesh J. Agril. Res. 33(3), 519-529.
- Reddy, M.A., Francies, R.M., Rasool, S.N. and Prakash-Reddy, V.R. (2014). Breeding for Tolerance to Stress Triggered by Salinity in Rice. International Journal of Applied Biology and Pharmaceutical Technology, 5, (1), 167-176.
- Reyes-Valde, M.H. (2000). A model for marker-based selection in gene introgression breeding programs. Crop Sci. 40, 91–98. https://doi.org/10.2135/cropsci2000.40191x
- Sahi, C., Singh, A., Kumar, K., Blumwald, E., and Grover, A. (2006). Salt stress response in rice: genetics, molecular biology, and comparative genomics. Functional Integrated Genomics, 6, 263–3. https://doi.org/10.1007/s10142-006-0032-5
- Sajjan, A.M., Lingaiah, H.B. and Fakrudin, B. (2016). Studies on genetic variability, heritability and genetic advance for yield and quality traits in Tomato (Solanum lycopersicum L.). International Journal of Horticulture, 6(18), 1-15. https://doi.org/10.5376/ijh.2016.06.0018
- Schwabele, K.A., Iddo, K. and Knap K.C. (2006). Drain water management for salinity mitigation in irrigated agriculture. American Journal of Agricultural Economics, 88, 133–140. https://doi.org/10.1111/j.1467-8276.2006.00843.x
- Shani, U. and Ben-Gal, A. (2005). Long-term response of grapevines to salinity: Osmotic effects and ion toxicity. Am. J. Enol. Vitic. 56, 148-154.
- Singh, R.K., Redona, E.D. and Refuerzo L. (2010). Varietal improvement for abiotic stress tolerance in crop plants: special reference to salinity in rice. In: Pareek, A., Sopory, S.K., Bohnert, H.J. and Govindjee (Eds.), Abiotic stress adaptation in plants: Physiological, Molecular and Genomic Foundation. Springer, New York, Pp. 387-415.
- Sivasubramanian, S. and Madhavamenon, P. (1973). Combining ability in rice. Madras Agricultural Journal, 60, 419-421.
- Sultana, J., Siddique, M. N. A. and Abdullah, M. R. (2015). Fertilizer recommendation for agriculture: practice, practicalities and adaptation in Bangladesh and Netherlands. International Journal of Business, Management and Social Research, 1(1), 21-40. https://doi.org/10.18801/ijbmsr.010115.03
- Summart, J., Thanonkeo, P., Panichajakul, S., Prathepha, P. and McManus, M.T. (2010). Effect of salt stress on growth, inorganic ion and proline accumulation in Thai aromatic rice, Khao Dawk Mali 105, callus culture. Afr. J. Biotechnol. 9, 145-152.
- Thomson, M.J., De Ocampo, M., Egdane, J., Rahman, M.A., Sajise, A.G., Adorada, D.L., Tumimbang-Raiz, E., Blumwald, E., Seraj, Z.I., Singh, R.K., Gregorio, G.B. and Ismail, A.M. (2010). Characterizing the SalTol quantitative trait locus for salinity tolerance in rice. Rice, 3, 148–160. https://doi.org/10.1007/s12284-010-9053-8
- Wassmann, R., Jagadish, S.V.K., Peng, S.B., Sumfleth, K., Hosen, Y., and Sander, B.O. (2010). Rice production and global climate change: scope for adaptation and mitigation activities. 67-76.
- Yeo, A.R., Yeo, M.E., Flowers, S.A. and Flowers, T.J. (1990). Screening of rice (Oryza sativa L.) genotypes for physiological characters contributing to salinity resistance, and their relationship to overall performance. Theor. Appl. Genet. 79, 377–384. https://doi.org/10.1007/BF01186082
- Yoshida, S., Forno, D.A., Cock, J.K. and Gomez, K.A. (1976). Laboratory manual for physiological studies of rice. Manila, Philippines: International Rice Research Institute.
- Zhou, W., Li, Y., Zhao, B.C., Ge, R.C., Shen, Y.Z., Wang, G. and Huang Z.J. (2009). Overexpression of TaSTRG gene improves salt and drought tolerance in rice. J. Plant Physiol. 166, 1660-1671. https://doi.org/10.1016/j.jplph.2009.04.015
© 2019 The Authors. This article is freely available for anyone to read, share, download, print, permitted for unrestricted use and build upon, provided that the original author(s) and publisher are given due credit. All Published articles are distributed under the Creative Commons Attribution 4.0 International License.
Journal of Bioscience and Agriculture Research EISSN 2312-7945.